Fickle Markers: Solutions For Antibody Binding Specificity Challenges

Reproducibility has been an ongoing, and important, concept in the sciences for years. In the area of biomedical research, the alarm was sounded by several papers published in the early 2010’s. Authors like Begley and Ellis, Prinz and coworkers, and Vasilevsky and colleagues, among others reported an alarming trend in the reproducibility of pre-clinical data. These reports indicated between 50% to almost 90% of published pre-clinical data were not reproducible. This was further highlighted in the article by Freedman and coworkers, who tried to identify and quantify the different sources of error that could be causing this crisis. Figure 1, taken from their paper, summarizes their efforts to quantify the different causes of error and the amount each contributes to the overall issue.

Even more concerning is the recent report from the Reproducibility Project: Cancer Biology, that attempted to reproduce the results from 53 cancer papers published in 2010-2012. Unfortunately, due to a variety of factors, only 23 papers were tested. The results stacked up to similar work, with only 43% of the experiments being reproducible; and more to the point, the size of the effect was much smaller than originally published.
As shown in Figure 1, about 36% of irreproducibility can be attributed to reagents and reference materials. In flow cytometry, our central reagent is the antibody so, the question is, how good are our reagents? In an article by Bradbury and Plückthun, they estimated that fewer than half of commercially available antibodies recognized only their specified target.
For flow cytometrists to do their part in the fight against poor reproducibility, we need to look at our reagents and evaluate how we characterize them before use. This article by Andersson et al., serves as a cautionary tale about antibody validation. For the past 20 years or so, researchers have been studying the estrogen receptor β (ERβ) with the hope of it being a breast cancer target. The authors performed a rigorous validation on the 13 available anti-ERβ antibodies and discovered that only one of them was specific to ERβ in immunochemistry. How much time, money, and effort was wasted because of poorly-validated reagents?
What Could Possibly Go Wrong With Your Antibody?
Let’s start by examining some of the issues that can occur with antibodies. After that, we will look at how to address these issues.
1. Lot-to-lot variability
In theory, hybridomas that produce monoclonal antibodies produce the identical antibody each time. There are several factors at work that might cause different results between lots, or between vendors. However, this may not be the case, as shown by Bradbury and coworkers (2018). In this paper, the researchers examined 185 random hybridomas and found that 31.9% of these produced additional heavy and/or light chains that would impact the specificity of the antibody to the intended target.
The second involves the conjugation of the fluorochrome to the antibody. For example, Szabó and coworkers (2018) showed that, in the case of the two fluorochromes tested, conjugation decreased antibody affinity, while Shrestha et al., (2012) showed that different conjugation targets also had differential effects on antibody affinity.
2. Cross-reactivity
Monoclonal antibodies are supposed to bind only to the target protein. In some cases, when too much antibody is used in an assay, non-specific binding can occur as the antibody may bind to an off-target protein. In other cases, the antibody might bind to a different antigen entirely.
In addition to the study mentioned above by Andersson et al., there are others studies that should give researchers pause when using antibodies. For example, Egelhofer and coworkers (2011) tested over 200 antibodies to different histone modifications and found that about one-quarterfailed specificity testing by dot or western blotting. Another article by Michel et al. (2009) looked at 46 antibodies against various G-coupled receptor proteins and found that most of the tested antibodies bound to more than one protein.
Sometimes cross-reactivity can be good, especially in proteins conserved through multiple species, allowing researchers to have access to antibodies against their target organism.
3. Unusual or unexpected binding patterns
Unusual binding patterns can be a cause for alarm. While these patterns may result in a new discovery, it is equally possible that they are the result of cross-reactivity with an off-target protein. A neglected issue is the binding of fluorochromes to cells. The first report of this was by van Vugt et al, (1996). More recently, Kirstensen and coworkers (2020) showed how cyanine dyes can bind non-specifically to monocytes. In an another interesting article by Hughes et al., (2020), the authors showed that the expression of PD-L1, a programmed death receptor, increased on neutrophils when incubated with AlexaFluor™700 fluorophore conjugated with anti-CD16, resulting in an artificial increase in PD-L1 signal in experiments.
4. Proper use
When using antibodies, it’s important that they are used for the correct assay. Depending on how the antibody is generated, it may recognize the native (folded) or denatured (linear) form of the protein. Likewise, the conditions in which the antibody is used can have an impact on its ability to bind the target. Thus, it is important to check with the vendor to see what applications the antibody has been tested in. If it is not listed, expect to do a lot of work to validate the antibody.
How To Validate Antibody Binding Specificity
Now that we know some of the issues that can arise with antibodies, let’s review some tools and techniques you can use to validate your reagents .
1. Turn to the literature
A good first step is to look into the literature; see what clones and vendors researchers commonly use. Check the OMIPs for newly designed and optimized multicolor panels. Other sites I find useful in identifying good antibodies include:
- Benchsci – a site that uses AI to analyze and characterize data in papers so that when a specific reagent is searched, the number of papers citing that antibody can be identified, along with figures from the paper.
- Antybuddy – a site for Amazon-like reviews of products, in particular antibodies
- Antibody Registry – a site that allows you to link your antibody to the vendor, has notes on applications as well.
- CiteAb – you can search for your antibody of interest and see citations from the literature.
If you know a good resource, please share it so we can get it up for everyone to enjoy.
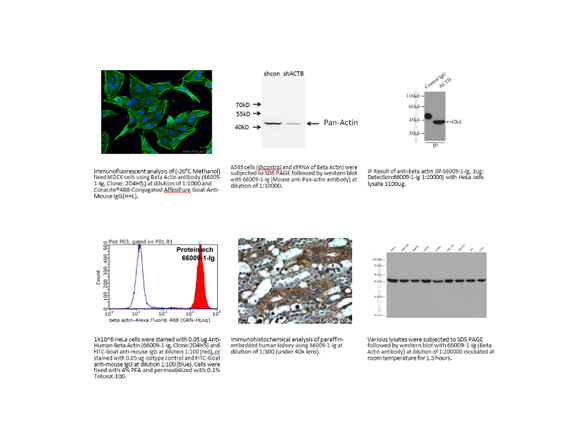
2. Vendor data
Don’t neglect the vendor’s data. Some vendors are better than others at sharing their validation data. Take this example in Figure 2 where a vendor shows the methods they use to validate an antibody.
Not every vendor provides as complete data as this, but this does add a level of confidence in the reagent.
3. Options for validation
You can always take the time to validate your reagent yourself. This option is the most time consuming, but in the end, you will know that your antibody is doing what it should be doing. One major limitation in validation is the motivation to do the work. Most of us purchase reagents assuming they will work, but it’s better to prove it. Here are some examples of how to validate an antibody.
- Positive and negative cell lines – with the advances in siRNA targeting, a positive cell line can be treated to knock down the expression of the target protein. Likewise, it is possible to transfect a negative cell to express the target protein. In both cases, there is a positive and a negative, thus showing if there is specific binding or not. Of course, you’ll have to prove by some other technique, such as PCR or mass spec, that the gene is either there or knocked down.
- Different techniques – using a different technique can also help confirm the specificity of the antibody. Immunoprecipitation is a great option, especially if coupled with mass spec analysis.

There are many more ways to test binding specificity. For example, this chart from Uhllen et al.(2016), summarizes different techniques, as well as what application the validation method is useful for.
I would also recommend reading this paper by Kalina et al. (2020) which summarizes these ideas while expanding on a few others.
4. Documentation
It seems intuitive that we should be writing things down – that’s what the lab notebook is for, right? That is true, but when you perform validation of reagents, it is best to make sure it is in a separate dedicated location for easy access.
Your report should contain all the information you used to confirm the reagent. Information from the vendor you acquired the antibody from, the lot specific information, titration results and more. This becomes your proof that the reagent worked as expected. It also helps when changing to a new lot of antibodies. You can confirm that the new reagent works equivalently to the original one. Should anyone have questions on the reagent, you can pull out the ‘book’ and have everything you need at your fingertips.
Conclusion
Antibodies are a critical reagent in flow cytometry experiments. They serve to identify the target cells of interest in a heterogeneous mix of cells, and they need to be fully validated to ensure that the results are robust and reproducible. In this way, we can avoid going down the rabbit hole. We can take a lesson from the chase to understand ERβ which led to 20 years of lost research opportunities. By building a culture of validation within the flow cytometry community, we can work to ensure that the data generated will be robust and reproducible.
To learn more about important control measures for your flow cytometry lab, and to get access to all of our advanced materials including 20 training videos, presentations, workbooks, and private group membership, get on the Flow Cytometry Mastery Class wait list.

ABOUT TIM BUSHNELL, PHD
Tim Bushnell holds a PhD in Biology from the Rensselaer Polytechnic Institute. He is a co-founder of—and didactic mind behind—ExCyte, the world’s leading flow cytometry training company, which organization boasts a veritable library of in-the-lab resources on sequencing, microscopy, and related topics in the life sciences.
More Written by Tim Bushnell, PhD












