The 5 Fundamental Methods For Imaging Nucleic Acids

A lot of microscopy assays are focused on labeling and imaging proteins. We often use antibodies against specific protein antigens or fuse a fluorescent protein to a protein of interest. These methods cover many applications, but maybe you are interested in viral RNA, gene duplication, or need a counterstain to label the nucleus. You can’t fuse a protein to nucleic acids, so where should you start?
Try these 5 assays to image nucleic acids:
Nucleic Acid Dyes
Nucleic acid dyes are small molecules that increase fluorescence when bound to nucleic acids. They can bind DNA, RNA, or both, but do not differentiate specific sequences.
1. DNA specific dyes
The two most well-known DNA specific dyes are Hoechst and DAPI. Both dyes bind the A-T rich regions in double-stranded DNA and are excited in the UV wavelengths. Most researchers use these dyes to label the nuclei of cells, but they can also be used for metaphase spread to karyotype chromosomal abnormalities.
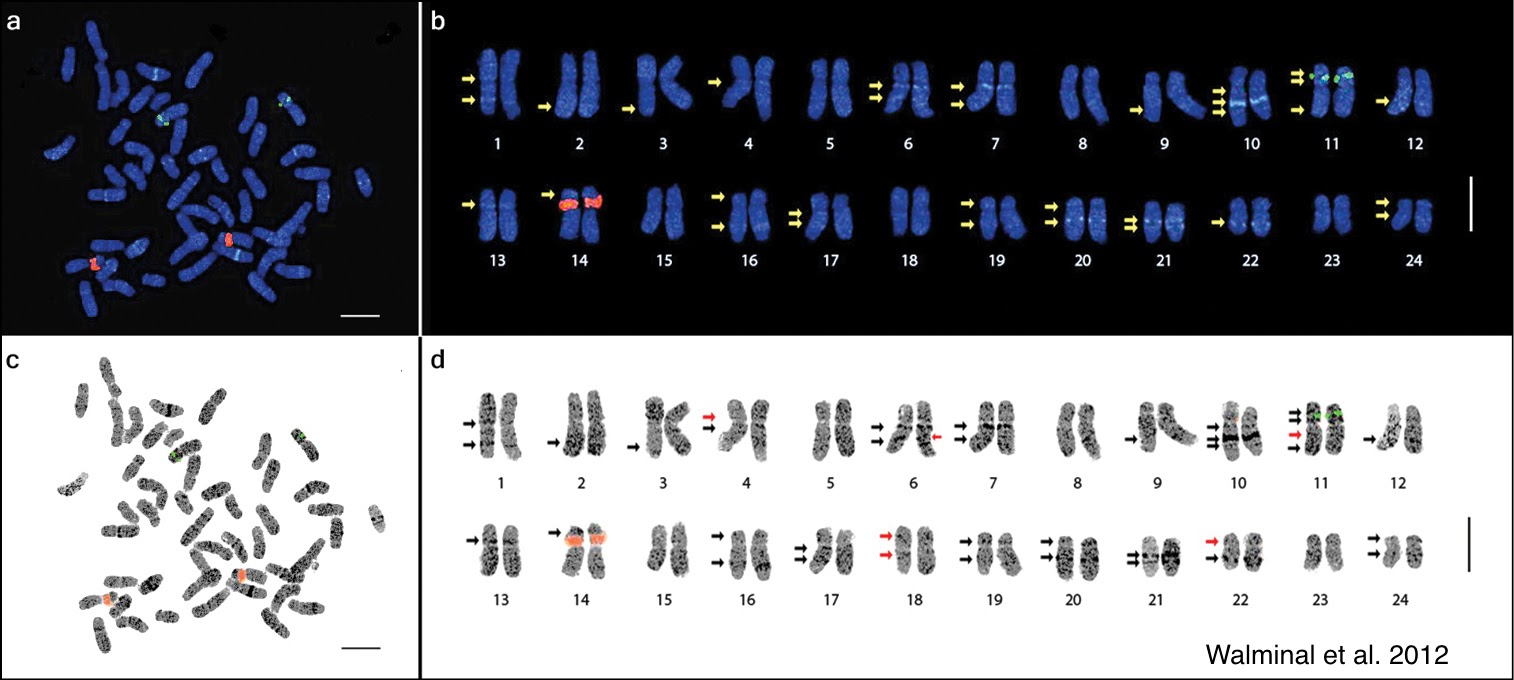
Fig. 1 Metaphase spread of Panax ginseng 2n=48 chromosomes (a and c) and the karyotype idiogram showing 24 homologous pairs (enlarged; b and d) arranged in decreasing lengths.
More recently, new DNA probes have been developed for use in the far red channel. DRAQ5 is a cell-permeable dye that binds dsDNA. The excitation for DRAQ 5 is 647nm, and its emission is 681nm. This red-shifted dye is an excellent choice for live-cell imaging applications because the UV radiation damages cells. Other DNA specific dyes released in the last few years are SiR-DNA and SPY-DNA from Spirochrome. The wide variety of dyes now allows imaging on many different specialty microscope techniques such as SIM and STED.
2. RNA specific dyes
There are less RNA specific dyes than DNA dyes, but one good option is SYTO RNAselect, which can be imaged in the GFP spectra when the dye is bound to RNA. SYTO RNAselect is currently the only commercially available RNA dye that can be used in live cells. Still, several publications on newly designed RNA dyes have emerged in the past few years and will likely become commercially available in the near future. RNAselect dye has been used to study RNA granules, ribosomal RNA (rRNA), exosomes carrying RNA, and viral replication cycle. These dyes have many applications that you have not even thought of before!

Fig. 2 Methanol-fixed MRC-5 cells stained with SYTO® RNASelect™ green-fluorescent cell stain.
3. General nucleic acid dyes
General nucleic acid dyes bind both RNA and DNA. These dyes may not be as specific but often make up for the lack of specificity with other desirable characteristics. One such dye is acridine orange, which can differentiate RNA and DNA based on the emission spectrum. When bound to dsDNA, acridine orange has a green emission (525nm), but when bound to RNA the dye has a red emission (650nm). This property makes the dye useful for comparing RNA and DNA content within a cell or during cell cycle progression.
The final general nucleic acid dye to know about is Propidium Iodide (PI). PI is most commonly used in flow cytometry for live/dead staining. This is because this dye can only permeate dying cells that have lost membrane integrity. In microscopy, PI it is used for cell health assays. One of the most common assays is using PI to determine the apoptosis of cells due to treatment.

Fig 3. Confocal laser scanning microscopy (CSLM) images of 24 h E. coli biofilm co-stained with propidium iodide (PI) and SYTO 9: vertical and horizontal cross-sections in multichannel
4. DNA Synthesis Labeling
We can use synthetic nucleotides to label cells that are proliferating and replicating their DNA. The two most common precursors for this technique are BrdU ( 5-bromo-2′-deoxyuridine) and EdU (5-ethynyl-2′-deoxyuridine). BrdU and Edu are non-radioactive methods of measuring DNA synthesis and have replaced the use of radioactive [3H]thymidine in many experiments. BrdU and EdU have some key differences to consider.
BrdU incorporation is followed by BrdU-specific antibody treatment during fluorescence microscopy. This means that the cell or tissue must be fixed before analysis, and it is not compatible with live-cell studies due to the size of the antibodies. BrdU has been combined with cell-specific markers to perform lineage analysis and cell fate studies. The one major limitation when using BrdU in experiments with tissue sections is that antibodies won’t penetrate well and will likely result in an undercount of actively cycling cells.
EdU is structurally similar to BrdU, but it has a terminal alkyne group. This alkyne group provides fluorescent labeling through “click” chemistry, which allows a fluorescent dye-conjugated azide to bind the target alkyne group through a covalent bond. EdU combined with “click” chemistry, which labels with a small molecule instead of an antibody, has vastly better penetration into tissue sections than BrdU.
Both BrdU and EdU have one major drawback for cell cycle or proliferation studies. Only cells that progress through S-phase and synthesize DNA during the treatment can incorporate the compounds. Compared to cells labeled with Ki67 (cell proliferation marker), BrdU and EdU undercount the number of actively proliferating cells. It is worthwhile to perform a comparison with your samples to ensure that your counts will be accurate.

Fig. 4 HeLa cells were fixed in 4% paraformaldehyde at RT for 15 min. Green: BrdU stained by BrdU antibody (ab152095) diluted at 1/2000. Blue: Hoechst 33342 staining.
5. In-situ hybridization
In-situ hybridization (ISH) is a method to label specific sequences of DNA or RNA and can be divided into chromogenic (CISH) and fluorescent (FISH) techniques. CISH and FISH only differ in how the probe is labeled and detected. CISH probes are generally labeled with horseradish peroxidase (HRP) or alkaline phosphatase (AP), while FISH probes are labeled with fluorochromes.
The methods for all versions of in-situ hybridizations are essentially the same. First, the sample is fixed and the DNA or RNA is denatured. Proper denaturing of the nucleic acid is critical because the complementary strand needs access to the target sequence. Next, the probe needs to bind the target sequence (hybridize) overnight. Finally, the sample needs to be blocked, and the probe detected.
There has been a considerable variety of methods using in situ hybridization in the last decade. Some versions that warrant further reading are:
- Single-molecule FISH (smFISH)
- NEAT RNA-FISH for lncRNA
- Multiplex-FISH (M-FISH/Chromosome Painting)
- COMET-FISH for DNA damage
- CARD-FISH for identifying microbes that can’t be cultured in the lab

Fig. 5 Schematic from “Combined In Situ Hybridization and Immunohistochemistry in Rat Brain Tissue Using Digoxigenin-Labeled Riboprobes.”
Although there is no current method to directly fuse RNA onto a protein or antibody to nucleic acid sequences, the scientific community has come up with innovative ways to image nucleic acids. Often we focus only on proteins within the cell. Still, it is becoming more evident that the localization of DNA or RNA within cells is a powerful tool for studying cell biology.
To learn more about important techniques for your flow cytometry lab, and to get access to all of our advanced materials including 20 training videos, presentations, workbooks, and private group membership, get on the Flow Cytometry Mastery Class wait list.

ABOUT HEATHER BROWN-HARDING
Heather Brown-Harding, PhD, is currently the assistant director of Wake Forest Microscopy and graduate teaching faculty.She also maintains a small research group that works on imaging of host-pathogen interactions. Heather is passionate about making science accessible to everyone.High-quality research shouldn’t be exclusive to elite institutions or made incomprehensible by unnecessary jargon. She created the modules for Excite Microscopy with this mission.
In her free time, she enjoys playing with her cat & dog, trying out new craft ciders and painting.You can find her on twitter (@microscopyEd) a few times of day discussing new imaging techniques with peers.
More Written by Heather Brown-Harding












