Critical Steps in DNA Cell Cycle Analysis

DNA cell cycle analysis is a very powerful technique in flow cytometry. It is deceptively easy, but there are several critical things to remember to ensure successful analysis.
Collect enough events.
Cell cycle analysis involves fitting of the data using one of several mathematical models that describe the behavior of the data. These models make different assumptions about the S phase as well as the G1 and G2/M phases. To have enough data, one should collect 100 events for each channel between the beginning of the G1 peak and the end of the G2/M peak. Thus, if the G1 peak starts at 40,000 and the G2/M phase ends at 110,000, the dataset should contain (110,000-40,000)*100 = 7,000,000 events.
Remember the stoichiometry.
The concentration of the DNA dye must be sufficient so that it binds in proportion to the amount of the DNA in the cell. Thus it is essential to have a good cell count to ensure the correct amount of DNA dye is added to the sample.
Fixation choice is important.
Crosslinking agents like formaldehyde will lower the dye binding because they introduce chromatin crosslinking. Dehydrating fixatives like methanol and ethanol are better, but at high concentration can cause cell clumping. Dehydrating dyes can also negatively impact fluorescent dyes if the DNA is being stained in association with surface maker stains. Don’t forget, a little detergent can help improve the access of the DNA dye.
Don’t forget the RNA.
Some dyes (PI, for example) will bind to both DNA and RNA. If using PI, it is critical to add an RNAse to the staining buffer. Failure to do so will result in messy DNA histograms.
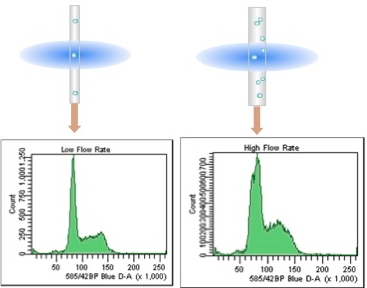
Watch the CV.
The CV of the G0/G1 peak is a measure of the quality of the DNA histogram. This can be affected by flow rate and laser alignment. The lower the CV the better, so it is critical to run DNA samples at low flow rates (narrow core streams) on a well aligned instrument.
Beware the doublets.
Doublets can masquerade as cells in the G2/M phase. It is critical to have good single cell prep for DNA cell cycle analysis. Watch the fixation steps and remember to filter the sample before running on the cytometer. Make sure to collect the pulse geometry measurements (H, W and A) to ensure that doublet discrimination gating can be performed on the sample.
Control the cell cycle.
It is a good practice to include a DNA cell cycle control into all experiments. Doing this allows for better characterization of changes in DNA cell cycle over time, allows for comparisons between samples/machines/days thus improving reproducibility and confidence. The most common are chicken RBC and trout RBC.
In summary, cell cycle analysis is a powerful tool in the flow cytometrists toolbox, but there are many optimization steps necessary for this deceptively easy assay. Don’t assume that one can add some PI to a sample and get a good DNA histogram. Choosing the best fixative for the assay, the right dye and a well behaved instrument are all critical for successful DNA cell cycle analysis.

ABOUT TIM BUSHNELL, PHD
Tim Bushnell holds a PhD in Biology from the Rensselaer Polytechnic Institute. He is a co-founder of—and didactic mind behind—ExCyte, the world’s leading flow cytometry training company, which organization boasts a veritable library of in-the-lab resources on sequencing, microscopy, and related topics in the life sciences.
More Written by Tim Bushnell, PhD