How To Use Flow Cytometry To Correctly Define T Cell Subsets And Their Functions

Flow Cytometry is a remarkably powerful tool for the study of T cells. It has been successfully used for many decades to accurately visualize and enumerate a variety of T cell subsets.
With a large sensitivity range for fluorescent probes, >95% sampling efficiency, and the ability to sort populations of interest for further study, fluorescent-based cytometry remains a tool of choice for T cell analysis.
Single cell visualization of T cells in a heterogeneous sample is clearest when the defined T cell populations are determined with ‘rock-solid’ gating and data analysis strategies.
For example, detection of the total CD4 and CD8 T cell compartments (via CD3+ CD4+ and CD3+ CD8+ cells, respectively) is straightforward; also, T cell populations that are clearly defined by surface antigen expression include antigen-specific (tetramer-binding) memory T cell clones and invariant Natural Killer T (iNKT) cells, a unique T cell subset discerned via binding to a CD1d-glycolipid loaded tetramer.
Such gating strategies, when paired with CD3 inclusion, doublet exclusion, and appropriate live/dead gating, allow clear, accurate visualization of your T cell population of interest and enumeration of frequency in your sample.
The Benefits And Caveats Of Advanced T Cell Antibody Panels
Flow panel sizes have expanded dramatically in the last 15+ years and continue to do so. An antibody panel with more than 10-colors is no longer uncommon, and with the adaptation of the more recent Brilliant dyes, 14+ colors are now very feasible on many instruments. With this increase in parameter detection per sample, the subsetting of T cells in the literature has exploded.
For example, when considering one aspect of T cell biology, the naïve to memory differentiation post-antigen exposure, the field has transcended from an era of two subsets (naïve and memory) into the era of Central Memory, Transitional Memory, Effector Memory, Terminally Differentiated Effector Memory, etc.
At first glance, these larger panels bring clarity to the T cell landscape. By revealing more of the complex marker distribution on individual cells, we gain a clearer picture of the heterogeneity of this facet of the immune cell compartment as a whole.
This, in turn, could allow better understanding of how the composite immune system functions in health and disease states (know your players, know the game) and also facilitate discovery of new therapeutic targets (á la anti-PD-1).
However, forcing square pegs into the round holes present in many linear differentiation models (with stages in the process noted via marker changes) can serve to further cloud the T cell field, potentially leading to misleading claims about the actual status of a T cell in a given sample or group of patients.
Some markers can change like the wind (it seems) and we must use caution to not underestimate the complex web of factors beyond mere ‘differentiation’ that impacts a T cell, causing it to express a given marker at a moment in time.
Why T Cell Function Should Guide Your T Cell Analysis
When trolling these waters, it’s best to pair our T cell subset findings with functional profiling of the population of interest.
The winds of T cell differentiation ‘states’ or who is or is not a bona-fide Treg may change, but a functional profile will serve to anchor the biological relevance and potential role of your T cell population of interest on more solid ground.
This is where flow cytometry cell sorting is advantageous, for sorting and functionally profiling T cell subsets (via a series of elispots or multiplex supernatant analysis of ex vivo cultures, for example) allows highly sensitive, multi-analyte profiling with far fewer cells than would be required for intracellular cytokine staining.
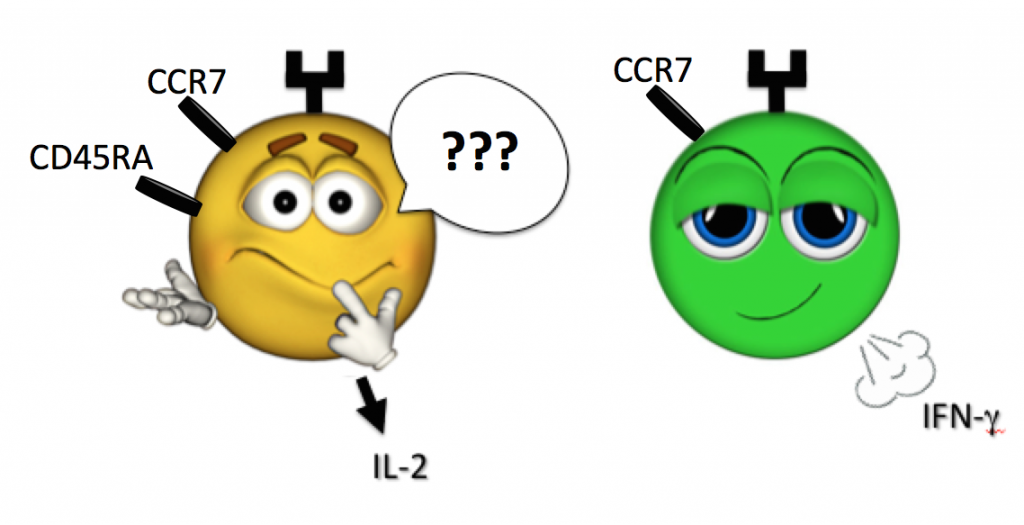
And what is a cell whose face certainly looks naïve (CD45RA+, CCR7+) yet is secreting IFN-g and TNF-a (as such cells have been found in human samples)?
When in doubt, let a cell’s actions lead its definition, and step back from pigeonholing via previously defined label. For example, defining a population as bearing a “surface phenotype resembling a central memory T cell, while secreting Th1 cytokines” is a safe and accurate way to go (see above Figure).
Overall, flow cytometry is an ideal way to visualize T cells in a heterogeneous sample. The key is to define your T cell populations of interest with correct gating strategies and to back up your T cell subset findings with functional analysis of these subsets. A cell’s actions should guide its definition, not the other way around.
To learn more about analyzing T cell subsets and subset function by flow cytometry, and to get access to all of our advanced materials including 20 training videos, presentations, workbooks, and private group membership, get on the Flow Cytometry Mastery Class wait list.

ABOUT TIM BUSHNELL, PHD
Tim Bushnell holds a PhD in Biology from the Rensselaer Polytechnic Institute. He is a co-founder of—and didactic mind behind—ExCyte, the world’s leading flow cytometry training company, which organization boasts a veritable library of in-the-lab resources on sequencing, microscopy, and related topics in the life sciences.
More Written by Tim Bushnell, PhD