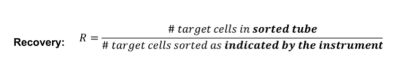Why Recovery, NOT Purity, Is The Best Measure Of A Cell Sorting Performance

One of the best perks of being a part of the flow cytometry community is the opportunity to attend one of the many regional flow meetings.
The chance to network with colleagues that do what you do is incredibly energizing.
It’s also a great opportunity to see what new research ideas they’re involved in before they’re published, and to talk with them directly about their ideas, and the research they’re involved in. I was recently at the Australasian Cytometry meeting and had a chance to talk to Dr. Rui Gardner, the Director of Flow Cytometry at the Gulbenkian Institute of Science in Lisbon, Portugal.
Rui gave a great talk on the concept of Rmax, which I’ll be talking about here. I want to make sure and acknowledge the other colleagues also involved in this work, who are Andy Riddell, Flow Cytometry Manager at Wellcome Trust-MRC Stem Cell Institute, Centre for Stem Cell Research University of Cambridge; Alexis Perez Gonzalez, Manager Flow Cytometry Core Facility, EMBL in Heidelberg; and Lola Martinez, Head of the Flow Cytometry Facility at CNIO, Madrid, Spain.
You can find this procedure in more detail here: https://www.youtube.com/watch?v=CUwbyuej9bA
Measuring Cell Sorter Performance
There seems to be quite a bit of confusion about how to measure the performance of a cell sorter, so I want to begin by defining a few terms, and then I want to discuss some of the variables that affect sort performance.
As a core manager for 30 years, I went through quite a transformation, as I learned that the sorter’s performance was very dependent on the sample preparation.
This is quite difficult to explain to the customer, though, so hopefully we can clear up some misconceptions.
One of the most important metrics to customers is the sort purity.
Purity is defined as :
Unfortunately, purity is NOT a good measure of instrument performance.
The instrument can be performing well, and the purity will be good, and the instrument can NOT be performing well, and the purity will also be good.
If the drop delay is not calculated correctly, the purity can be good, but the recovery will suffer.
There have been a few publications and posters on this, but it has gone largely unnoticed that measuring purity is not the best way to measure sort performance or to calculate the drop delay, but it is the method that most commercial sorters use to do this.
How do we do a better job?
Calculating Recovery, Yield & Purity
If we want to calculate the actual yield, we have a lot of counting methods, like cell counters and Neubauer chambers, but these all come with a fairly high level of associated error.
The best method would be independent of this error.
As long as we sort similar particles, which show the same Poisson behavior when sorted, the following holds.
At any given time, after a fraction α of the original sample is sorted, the absolute number of target particles (t) in this fraction (α.Ot) is distributed between the sorted tube (St) and the center stream catch (Ct) collected during the sort:
The same applies for non-target particles where:
We can estimate the maximum recovery for a given set of sort conditions:
If sort purity approaches 100%, this simplifies to:
The first time I saw the above equation, I was thinking this was going to be a complicated experiment, but Rui and colleagues have given us all of the tools to make it really easy to do.
The experiment is outlined in the protocol I linked above.
Adjusting Drop Delay For Purity Versus Recovery
You set up a sort with two different particle types, and measure the proportions of each in the sort tube and the waste.
The data are entered into the spreadsheet provided, and the maximum recovery can be predicted.
By comparing the recovery and purity at the drop delay calculated by the instrument, and by sorting slightly above and below this value, you can see that the calculated drop delay is rarely the ideal for recovery.
As you can see from the graphs below, from two different instruments, the calculated delay works very well for purity, but for maximal recovery, the delay should be shifted.
In sum, recovery is much more sensitive to the correct calculation of the drop delay than the purity, and we should be sure we choose the correct metric for measuring sort performance.
To learn more about cell sorting and to get access to all of our advanced materials including 20 training videos, presentations, workbooks, and private group membership, get on Mastery Class wait list.

ABOUT TIM BUSHNELL, PHD
Tim Bushnell holds a PhD in Biology from the Rensselaer Polytechnic Institute. He is a co-founder of—and didactic mind behind—ExCyte, the world’s leading flow cytometry training company, which organization boasts a veritable library of in-the-lab resources on sequencing, microscopy, and related topics in the life sciences.
More Written by Tim Bushnell, PhD